active site
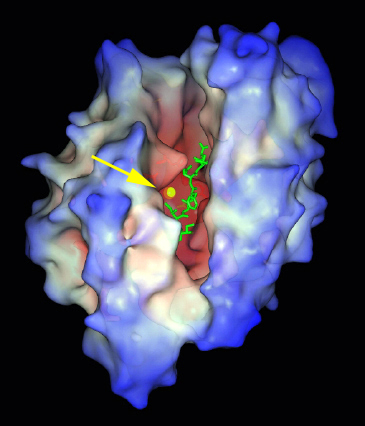
A computer model of the active site of an enzyme called mammalian zinc metalloendopeptidase, or EP 24.15. Credit: M. J. Glucksman.
An active site is a region on the surface of an enzyme to which a specific substrate or set of substrates binds. The properties of the active site are determined by the sequences of amino acids and the three-dimensional arrangement of the polypeptide chains of the enzyme.
The active site is not a point or plane but an intricate pocket or cleft – a 3-dimensional entity – structurally tailored to accept a particular substrate or group of substrates. The cleft or crevice excludes water from the active site, which would disrupt bond-breaking and -making processes, and is surrounded by non-polar amino acid residues, which give the active site a non-polar environment, important for both binding and catalysis.
Enzyme-substrate complex
The specific action of an enzyme with a single substrate can be partly understood using the lock and key analogy first proposed by Emil Fischer in 1894. In this analogy, the lock is the enzyme and the key is the substrate. Only the correctly sized and shaped key (substrate) fits into the key hole (active site) of the lock (enzyme).
However, not all experimental data can be adequately explained by using the so-called rigid enzyme model assumed by the lock and key theory. For this reason, a modification called the induced-fit theory has been proposed.
The induced-fit theory assumes that the substrate plays a role in determining the final shape of the enzyme and that the enzyme has some flexibility. This explains why certain compounds can bind to the enzyme but do not react because the enzyme has been distorted too much. Other molecules may be too small to induce the proper alignment and therefore cannot react. Only the proper substrate is capable of inducing the proper alignment of the active site.
The substrate binds to the enzyme by relatively weak forces to form the enzyme-substrate complex. The free energy of interaction between enzyme and substrate ranges from –12 to –36 kJ/mole, whereas the strength of a covalent bond can be up to –450 kJ/mole.


