DNA (deoxyribonucleic acid)
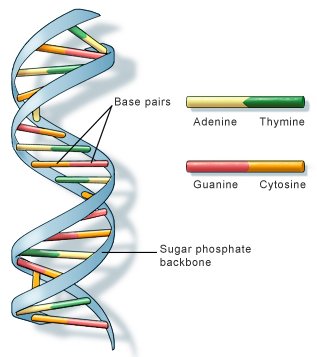
Fig 1. The double helix structure of a DNA molecule. US National Library of Medicine.
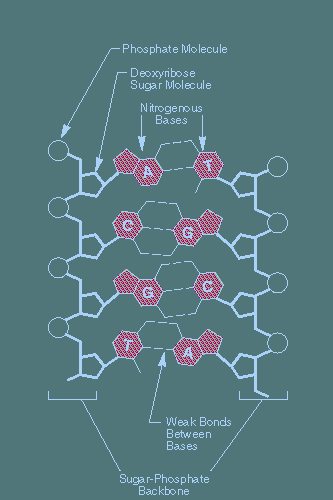
Fig 2. Main chemical components of a DNA molecule.
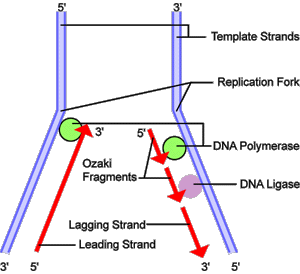
Fig 3. DNA replication. Before a cell can divide, it must first duplicate its DNA. This figure provides an overview of the DNA replication process. In the first step, a portion of the double helix (blue) is unwound by a helicase. Next, a molecule of DNA polymerase (green) binds to one strand of the DNA. It moves along the strand, using it as a template for assembling a leading strand (red) of nucleotides and reforming a double helix. Because DNA synthesis can only occur 5' to 3', a second DNA polymerase molecule (also green) is used to bind to the other template strand as the double helix opens. This molecule must synthesize discontinuous segments of polynucleotides (called Okazaki Fragments). Another enzyme, DNA Ligase (yellow), then stitches these together into the lagging strand.
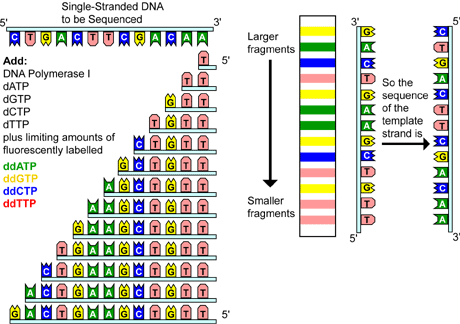
Fig 4. Chain termination sequencing involves the synthesis of new strands of DNA complementary to a single-stranded template (step I). The template DNA is supplied with a mixture of all four deoxynucleotides, four dideoxynucleotides (each labeled with a different colored fluorescent tag), and DNA polymerase (step II). Because all four deoxynucleotides are present, chain elongation proceeds until, by chance, DNA polymerase inserts a dideoxynucleotide. The result is a new set of DNA chains, all of different lengths (step III). The fragments are then separated by size using gel electrophoresis (step IV). As each labeled DNA fragment passes a detector at the bottom of the gel, the color is recorded. The DNA sequence is then reconstructed from the pattern of colors representing each nucleotide sequence (step V).
DNA (deoxyribonucleic acid) is the genetic material of most terrestrial organisms and one of two principal types of nucleic acid found in living cells (the other being RNA). DNA is a stable macromolecule consisting (usually) of two strands running in opposite directions. These strands twist around one another in the form of a double helix and are built up from components known as nucleotides. The double helix nature of DNA was established, among others, by Francis Crick, Rosalind Franklin, and James Watson.
There are 10 nucleotides per turn in the double helix (1 turn = 3.4 nanometer). Each nucleotide consists of a sugar (deoxyribose)-phosphate section attached to one of four organic bases – two purines, guanine and thymine, and two pyrimidines, adenine and cytosine. The bases on opposite strands project toward each other like the rungs of a twisted ladder.
Complimentary base pairing is highly specific: thymine always pairs with adenine (via 2 hydrogen bonds) and cytosine always pairs with guanine (via 3 hydrogen bonds). This specificity is a direct consequence of the shapes of the bases and is fundamental to terrestrial life since it unde#rlies all aspects of inheritance and gene expression including DNA replication, DNA repair, transcription, RNA splicing, and translation.
The sequence of bases along the length of a DNA strand varies from species to species and from individual to individual, and determines the organism's development (see genetic code) by controlling protein synthesis. Although double-stranded DNA forms the genetic material for most terrestrial organisms, bacteriophages and viruses may use single-stranded DNA, single-stranded RNA, or double-stranded RNA.
That DNA might be able to survive for hundreds of thousands of years in the vacuum of space is suggested by the results of research announced in 1998 by Evan Williams and his colleagues at the University of California, Berkeley.1 The Berkeley group concluded that double-stranded DNA could maintain its structure in a vacuum for as long as 35 years at room temperature, and perhaps almost indefinitely in the very low temperatures of space.
DNA hybridization
DNA hybridization is a method of comparing the DNA from different organisms in order to discover genetic relationships between species. Single strands of DNA from each species are placed together and allowed to react. Some strands will "hybridize" to form double strands (the usual structure of DNA) and the extent to which they do so indicates how many base sequences are complimentary. This in turn is a measure of how similar the genes are.
DNA repair
Maintenance of the accuracy of the DNA genetic code is critical for both the long- and short-term survival of cells and species. Sometimes, normal cellular activities, such as replicating DNA and making new gametes, introduce changes or mutations in our DNA. Other changes are caused by exposure of DNA to chemicals, radiation, or other adverse environmental conditions. No matter the source, genetic mutations have the potential for both positive and negative effects on an individual as well as its species. A positive change results in a slightly different version of a gene that might eventually prove beneficial in the face of a new disease or changing environmental conditions. Such beneficial changes are the cornerstone of evolution. Other mutations are considered deleterious, or result in damage to a cell or an individual. For example, errors within a particular DNA sequence may end up either preventing a vital protein from being made or encoding a defective protein. It is often these types of errors that lead to various disease states.
The potential for DNA damage is counteracted by a vigorous surveillance and repair system. Within this system, there are a number of enzymes capable of repairing damage to DNA. Some of these enzymes are specific for a particular type of damage, whereas others can handle a range of mutation types. These systems also differ in the degree to which they are able to restore the normal, or wild-type, sequence.
Categories of DNA repair systems
Photoreactivation is the process whereby genetic damage caused by ultraviolet radiation is reversed by subsequent illumination with visible or near-ultraviolet light.
Nucleotide excision repair is used to fix DNA lesions, such as single-stranded breaks or damaged bases, and occurs in stages. The first stage involves recognition of the damaged region. In the second stage, two enzymatic reactions serve to remove, or excise, the damaged sequence. The third stage involves synthesis by DNA polymerase of the excised nucleotides using the second intact strand of DNA as a template. Lastly, DNA ligase joins the newly synthesized segment to the existing ends of the originally damaged DNA strand.
Recombination repair, or post-replication repair, fixes DNA damage by a strand exchange from the other daughter chromosome. Because it involves homologous recombination, it is largely error free.
Base excision repair allows for the identification and removal of wrong bases, typically attributable to deamination – the removal of an amino group (NH2) – of normal bases as well as from chemical modification.
Mismatch repair is a multi-enzyme system that recognizes inappropriately matched bases in DNA and replaces one of the two bases with one that "matches" the other. The major problem here is recognizing which of the mismatched bases is incorrect and therefore should be removed and replaced.
Adaptive/inducible repair describes several protein activities that recognize very specific modified bases. They then transfer this modifying group from the DNA to themselves, and, in doing so, destroy their own function. These proteins are referred to as inducible because they tend to regulate their own synthesis. For example, exposure to modifying agents induces, or turns on, more synthesis and therefore adaptation. SOS repair or inducible error-prone repair is a repair process that occurs in bacteria and is induced, or switched on, in the presence of potentially lethal stresses, such as UV irradiation or the inactivation of genes essential for replication. Some responses to this type of stress include mutagenesis – the production of mutations – or cell elongation without cell division. In this type of repair process, replication of the DNA template is extremely inaccurate. Obviously, such a repair system must be a desperate recourse for the cell, allowing replication past a region where the wild-type sequence has been lost.
DNA replication
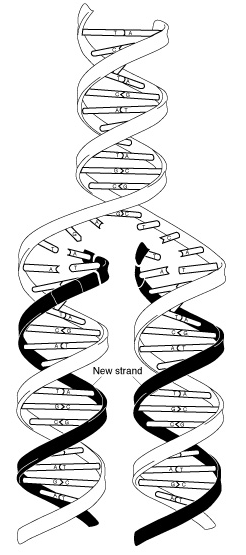 |
DNA replication (Fig 3) is the process of duplicating a cell's genome, required at each cell division. Replication, like all cellular activities, requires specialized proteinsfor carrying out the job. In the first step of replication, a special protein, called a helicase, unwinds a portion of the parental DNA double helix. Next, a molecule of DNA polymerase – a common name for two categories of enzymes that influence the synthesis of DNA – binds to one strand of the DNA. DNA polymerase begins to move along the DNA strand in the 3' to 5' direction, using the single-stranded DNA as a template. This newly synthesized strand is called the leading strand and is necessary for forming new nucleotides and reforming a double helix. Because DNA synthesis can only occur in the 5' to 3' direction, a second DNA polymerase molecule is used to bind to the other template strand as the double helix opens. This molecule synthesizes discontinuous segments of polynucleotides, called Okazaki fragments. Another enzyme, called DNA ligase, is responsible for stitching these fragments together into what is called the lagging strand.
The average human chromosome contains an enormous number of nucleotide pairs that are copied at about 50 base pairs per second. Yet, the entire replication process takes only about an hour. This is because there are many replication origin sites on a eukaryotic chromosome. Therefore, replication can begin at some origins earlier than at others. As replication nears completion, "bubbles" of newly replicated DNA meet and fuse, forming two new molecules.
With multiple replication origin sites, one might ask, how does the cell know which DNA has already been replicated and which still awaits replication? To date, two replication control mechanisms have been identified: one positive and one negative. For DNA to be replicated, each replication origin site must be bound by a set of proteins called the Origin Recognition Complex. These remain attached to the DNA throughout the replication process. Specific accessory proteins, called licensing factors, must also be present for initiation of replication. Destruction of these proteins after initiation of replication prevents further replication cycles from occurring. This is because licensing factors are only produced when the nuclear membrane of a cell breaks down during mitosis.
DNA sequencing
DNA sequencing is determining the exact order of the base pairs in a segment of DNA. In 1977, 24 years after the discovery of the structure of DNA, two separate methods for sequencing DNA were developed: the chain termination method and the chemical degradation method. Both methods were equally popular to begin with, but, for many reasons, the chain termination method is the method more commonly used today. This method is based on the principle that single-stranded DNA molecules that differ in length by just a single nucleotide can be separated from one another using a technique called polyacrylamide gel electrophoresis.
The DNA to be sequenced, called the template DNA, is first prepared as a single-stranded DNA. Next, a short oligonucleotide is annealed, or joined, to the same position on each template strand. The oligonucleotide acts as a primer for the synthesis of a new DNA strand that will be complementary to the template DNA. This technique requires that four nucleotide-specific reactions – one each for G (guanine), A (adenine), C (cytosine), and T (thymine) – be performed on four identical samples of DNA. The four sequencing reactions require the addition of all the components necessary to synthesize and label new DNA, including:
A DNA template
A primer tagged with a mildly radioactive molecule or a light-emitting
chemical
DNA polymerase, an enzyme that drives the synthesis of DNA
Four deoxynucleotides (G, A, C, and T)
One dideoxynucleotide, either ddG, ddA, ddC, or ddT
After the first deoxynucleotide is added to the growing complementary sequence, DNA polymerase moves along the template and continues to add base after base. The strand synthesis reaction continues until a dideoxynucleotide is added, blocking further elongation. This is because dideoxynucleotides are missing a special group of molecules, called a 3'-hydroxyl group, needed to form a connection with the next nucleotide. Only a small amount of a dideoxynucleotide is added to each reaction, allowing different reactions to proceed for various lengths of time until by chance, DNA polymerase inserts a dideoxynucleotide, terminating the reaction. Therefore, the result is a set of new chains, all of different lengths.
To read the newly generated sequence, the four reactions are run side-by-side on a polyacrylamide sequencing gel. The family of molecules generated in the presence of ddATP is loaded into one lane of the gel, and the other three families, generated with ddCTP, ddGTP, and ddTTP, are loaded into three adjacent lanes. After electrophoresis, the DNA sequence can be read directly from the positions of the bands in the gel.
Variations of this method have been developed for automated sequencing machines. In one method, called cycle sequencing, the dideoxynucleotides, not the primers, are tagged with different colored fluorescent dyes; thus, all four reactions occur in the same tube and are separated in the same lane on the gel. As each labeled DNA fragment passes a detector at the bottom of the gel, the color is recorded, and the sequence is reconstructed from the pattern of colors representing each nucleotide in the sequence.
Researchers can use DNA sequencing to search for genetic variations and/or mutations that may play a role in the development or progression of a disease. The disease-causing change may be as small as the substitution, deletion, or addition of a single base pair or as large as a deletion of thousands of bases.
Recombinant DNA
Recombinant DNA consists of hybrid molecules made by isolating segments of DNA and splicing them together with other DNA fragments to create molecules with specific, novel properties. Recombinant DNA is used by molecular biologists to study the expression of genes.
Reference
1. Schnier, P. D., Klassen, J. S., Strittmatter, E. F., and Williams, E. R. "Activation Energies for Dissociation of Double Strand Oligonucleotide Anions: Evidence for Watson-Crick Base Pairing in Vacuo," Journal of the American Chemical Society, 120 (37), 9605 (1998).


